Machine-Readable Chemical Structures
Introduction
Finding relevant articles based on IUPAC names or trivial names of molecules is a challenging task, while chemical identifiers allow for unambiguous identification of compounds. Redrawing of chemical structures is labour and time intensive, while chemical table (CT) files or SMILES structure codes can be used without any additional effort with any common structure drawing software.
Having machine-readable chemical structures as CT files, such as mol files, and as SMILES structure codes plus the InChI identifiers as part of a dataset associated with a research article will enhance its findability by making the article easily indexable and structure searchable. This also improves interoperability and facilitates reuse of scientific work.
While such information is not required for structured field-specific repositories such as Chemotion Repository, as this information is provided by the repository software, datasets in generic repositories benefit from this information.
The following will provide a tutorial on how to increase the machine-readability of chemistry research articles by providing machine-readable chemical structures within the associated dataset. Recommendations to provide structure codes and identifiers in a machine-readable supplementary table within the associated dataset are given.
Get mol files
All common structure drawing software save mol files. Copy the structure to a new document within your preferred structure drawing software. Then, choose File -> Save As -> choose MDL Molfile from the dropdown menu -> Save.
The name of the file might be chosen following your lab journal entry and also numbering of the structure in the article.
Get SMILES, InChI and InChIKey
ChemDoodle
To retrieve SMILES, InChI and InChIKey in ChemDoodle select a structure, then choose Edit -> Copy As -> Daylight SMILES or IUPAC InChI.
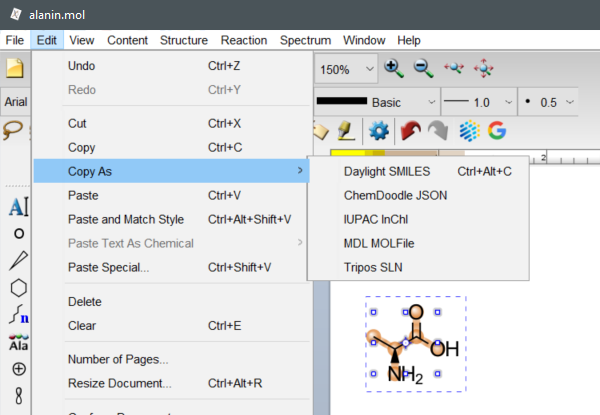
(ChemDoodle v11.7.0, iChemLabs, LCC., Chesterfield, VA, United States, 2021.)
You may also select a structure, then choose Structure -> Generate Line Notation -> Daylight SMILES or IUPAC InChI.
Alternatively, SMILES, InChI and InChIKey can also be saved as text files by choosing File -> Save as -> choose Daylight SMILES or InChI from the dropdown menu -> Save.
Choosing “IUPAC InChI” will also provide the InChIKey if enabled under Preferences. To include InChIKey, choose Edit -> Preferences -> Files tab -> scroll down and tick Include InChI key.
ChemDraw Professional
To retrieve SMILES, InChI and InChIKey in ChemDraw Professional, select a structure, then choose Edit -> Copy As -> SMILES, InChI or InChIKey.
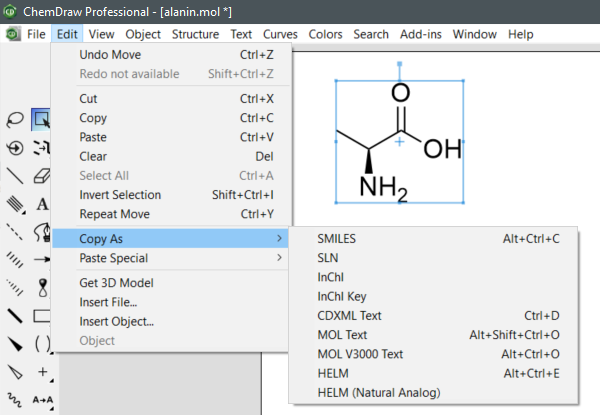 "
"
(ChemDraw Professional v20.1.0.11, PerkinElmer Informatics, Inc., Waltham, MA, United States, 2021.)
ChemSketch
To retrieve SMILES, InChI and InChIKey in ACD/ChemSketch, select a structure, then choose Tools-> Generate -> SMILES Notation or InChI for Structure.
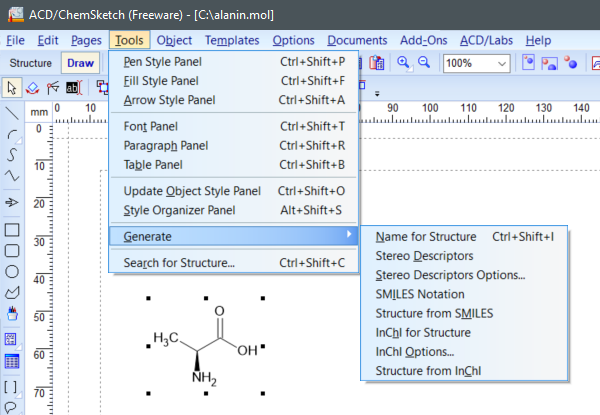
(ACD/ChemSketch v2021.1.1, Advanced Chemistry Development, Inc., Toronto, ON, Canada, 2021.)
Choosing InChI for Structure will also provide the InChIKey, if enabled under InChI Options. To include InChIKey, choose Tools -> Generate -> InChI Options and tick InChI Key.
MarvinSketch
To retrieve SMILES, InChI and InChIKey in MarvinSketch select a structure, then choose Edit -> Copy As. In a new windows choose Daylight SMILES, InChI/RInChI or InChIKey/RInChIKey.
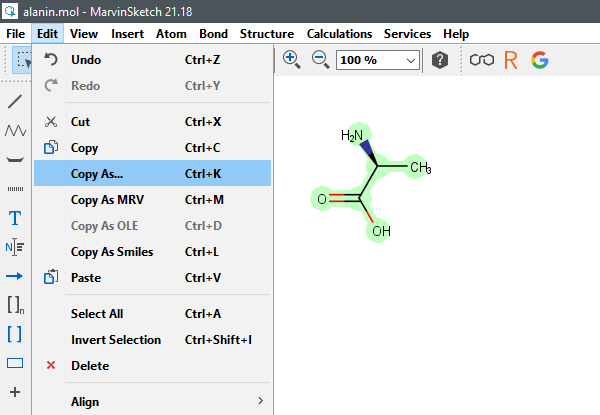
(MarvinSketch v21.18, ChemAxon, Ltd., Budapest, Hungary, 2021.)
Alternatively, SMILES, InChI and InChIKey can also be saved as text files by choosing File -> Save as -> choose Daylight SMILES, InChI/RInChI or InChIKey/RInChIKey from the dropdown menu -> Save.
What do you need and when?
CT files, SMILES and InChI are different representations of chemical structures. An important feature of CT files is the ability to store 3D data of molecules. CT files are the right choice for describing the 3D structure of molecules in a machine-readable way, e.g. obtained by using single-crystal X-ray diffraction (XRD). For all other use cases, InChI and SMILES are sufficient representations with the additional beauty that they are (simple) line notations.
Please note that InChI is an identifier, while SMILES is a structure code. Conversion of SMILES to a chemical structure graph and back to SMILES is possible, while this will not necessarily give the same SMILES as initially provided, hence, SMILES is not an identifier. On the other hand, InChI is an identifier and is not designed to be used to regenerate the correct chemical structure drawing, as it knows connectivities, not the bond order.
SMILES and InChI as well as InChI Key are ideal for describing chemical structures in research articles in a supplementary table. Such a table should be part of a dataset in a generic repository, but could also be submitted with the manuscript to the academic publisher, as required by ACS Journal of Medicinal Chemistry for SMILES since 2014.
Provide Machine-Readable Data as Supplementary Table
Information to enhance machine-readability might be provided within a dataset as a supplementary table, as there aren't open formats for all types of chemistry data available yet that would provide this information. This table should be provided in a machine-readable text-based format, such as a CSV file.
The minimum and recommended columns of such a table in chemistry are as follows:
- Letter-code and number in your lab journal, i.e. a local sample identifier
- Number(s) of structures in the article, i.e. a local structure identifier within your (corresponding) article
- SMILES
- InChI
- InChIKey
The letter-code and number in your lab journal should be included, as analytical data files in a dataset are frequently named following the lab journal entries.
Additionally, this table may also include further columns for:
- CAS registry number
- IUPAC name
- synonym i.e. common names of a compound
- PubChem compound identifier
- CAS registry number
- comment (if required)
Templates for such a table containing the minimum recommended columns are provided as .ods, .xlsx, and .csv files. These template files also take advantage of ontologies to unambiguously identify terms for humans and machines.
If the experimental work is documented in an ELN, these information could also be provided by the ELN system. Chemotion ELN generates SMILES, InChI, InChIKey as well as RInChI and RInChIKey for compounds and reactions. These information are also available in Chemotion Repository i.e. such a supplementary table is not required with structured, field-specific repositories such as Chemotion Repository, while datasets in generic repositories will profit from such a table.
This page is licensed under a Creative Commons Universal (CC0 1.0) Public Domain Dedication International Licence.
Main author: ORCID:0000-0003-4480-8661