Best Practices
On this page, we provide best practices for the publication of data in field-specific as well as generic research data repositories.
Best practices for field-specific repositories
For the field-specific repositories Chemotion Repository in combination with Chemotion ELN, the data is collected along the scientific workflows, analytical data files are automatically converted to open formats, controlled vocabularies or ontologies are used to describe data, and analysis data is interconnected to reactions and samples, hence, chemical structures. The entire Chemotion package allows researchers to collect, analyse, process, store, and publish various types of analytical data attached to reactions and samples in one digital environment. FAIRification of data along this data life cycle is mainly handled by Chemotion. During the seamless export from the ELN into the repository, PIDs (in this case DOIs) are assigned to the deposited data. Chemotion Repository provides DOIs for reactions, samples, analysis data, and collections (Collection DOI).
Other field-specific repositories such as CSD and ICSD, Strenda DB, SupraBank, NOMAD, and Chemotion Repository, if used independently from Chemotion ELN, offer built-in workflows for the aggregation of structured data along the submission pathways, aligned with the FAIR Data Principles. This greatly facilitates the publication of FAIR data.
Further aspects to consider are to provide rich metadata, which could be domain-independent metadata or domain-specific metadata, and should also include provenance information, which previously would have typically been included in the supplemental PDF section on general information and protocols. Minimum information requirements should be followed, as set by the chemistry community, and implemented in articles and previously also in supplementary PDFs.
Are you asking yourself right now, "That's it?" Yes, that's it! As mentioned above, field specific repositories carry out much of the heavy lifting when it comes to FAIRification, while a digital smart lab environment even collects structured data directly along the scientific workflows and data life cycle.
Best practice examples for field-specific repositories
For a better understanding of data publishing in field-specific repositories and how this data could be linked to related journal publications, the following examples including their data can be accessed via the following Lead by Example pages:
- in Chemotion Repository: Modular Synthesis of New Pyrroloquinoline Quinone Derivatives.
- in Chemotion Repository and CSD/CCDC: Modular Synthesis of trans‐A2B2‐Porphyrins with Terminal Esters: Systematically Extending the Scope of Linear Linkers for Porphyrin‐Based MOFs.
Best practices for generic repositories
While field-specific repositories provide significant support for the FAIRification of data along the entire data life cycle, the preparation of datasets for data publishing in generic repositories occurs at the disclosure and publication stage, as long as no (institutional) RDM workflows streamline data publishing. Usually, data is gathered simultaneously with the preparation of a corresponding manuscript, hence, datasets which are planned to be published with a generic repository require manual FAIRification prior upload and publishing.
To start, all data to be published should be collected and ordered in a logical folder structure, e.g. a folder for NMR and another folder for MS data (figure 1). Nested folder structures should be avoided and aspects of data organisation, such as file naming, should be taken into account. Researchers should aim for a data package which is as self-explanatory as the supplemental PDFs, they have previously published along scientific articles.
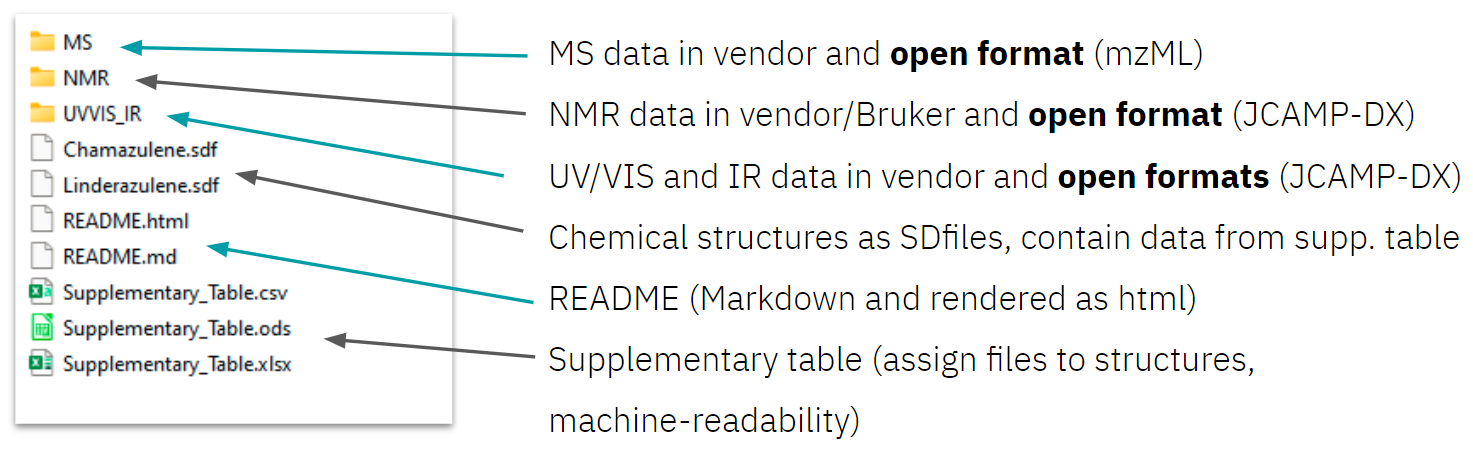
Figure 1: Example folder structure and content of the Lead by Example dataset of Linderazuelene.
For further aspects to consider, we provide a non-exhaustive list of generic best practices on what a chemistry dataset should contain and how the chemical data should be represented and described. Please note, that domain-specific aspects also need to be considered.
- Data should be published in open formats along the original raw data in proprietary formats.
Open formats for analytical data should be the main choice. However, many analytical instruments provide data in proprietary formats. Not all data in these formats is necessarily also included in the selected open format it was converted to, depending on the specification of the format applied. Hence, the original raw data (in proprietary formats) should also be published, although this data may have limited interoperability. Publishing original raw data is a measure of scientific integrity and allows for unbiased reprocessing and reuse of data. If no open format exists, export as text file, i.e. without any format specification, should be considered. - Data should be linked to chemical structures and reactions.
As analytical data are usually named following lab journal entries, the dataset must include a description of which data correspond to which sample, molecule, or experiment. This should be included in the format used for analytical data. Another solution on that is to provide a supplementary table (figure 1) within the data package, which should include InChI structure identifiers and SMILES structure codes and additional information such as RInChI reaction identifiers. Second-tier, chemical structures might also be added and represented as CT files such as mol or SDfiles (figure 1). RXNfiles may be used to describe chemical reactions in a machine readable way. - Datasets should include scripts and workflows and information on software used.
Workflows, i.e. scripts used for data processing, and input parameters for semi-automatic scripts, should be included. The metadata of the whole dataset should describe the language and version of scripts used as well as other software applied. Best practice is not only to include the code but to add notebooks such as R Notebooks or Jupyther notebooks. - Data should be described with metadata and minimum information standards should be followed.
Domain-independent metadata, via the repository's metadata editor, should be provided. Additionally, domain-specific metadata should be published via the metadata editor, if supported by the repository of choice, or within the analytical data or an additional README file. Minimum information requirements as set by the chemistry community and followed in manuscripts and previously also in supplementary PDFs should be included. - The provenance information of data should be included.
Part of that provenance information is part of the dataset's metadata and should be added via the metadata editor of the repository. All information which previously would have typically been included in the supplemental PDF section on general information and protocols, e.g. information on methods and instruments used, are provenance information and should be added to a README file, as long as no domain-specific metadata schema is available. This README could be a text file, or written in Markdown, while also a human-readable rendered representation as HTML could be provided.
Does that sound like a lot of manual work? Avoid extra work by using a smart lab digital environments for collecting, processing, analysing, and publishing research data, such as Chemotion ELN in combination with Chemotion Repository! Plus, you may omit the preparation of supplementary PDFs and use the saved work time to prepare your dataset for publication!
Best practice examples for generic repositories
For a better understanding of data publishing in generic repositories and how this data could be linked to related journal publications, the following examples, including their data, can be accessed via the following Lead by Example pages:
- in RADAR4Chem: Linderazulen aus einer invasiven Pflanze - Delphi und sein violettes Wunder.
- in Zenodo: A dataset of 255,000 randomly selected and manually classified extracted ion chromatograms for evaluation of peak detection methods.
- in RADAR: Synthesis and biological evaluation of highly potent fungicidal deoxy-hygrophorones.
- in DaRus: Predictive design of ordered mesoporous silica with well-defined, ultra-large mesopores.
Main authors: ORCID:0000-0003-4480-8661, ORCID:0000-0002-6243-2840 and ORCID:0000-0003-2060-842X